QHG Tutorial 08 - Simple Predator-Prey¶
In this example we model a simple predator-prey system, consisting of stationarsy agents (one per cell) representing grass, and moving agents representing sheep who like to eat grass.
Sheep and grass populatins communcate with each other by means of shared arrays.
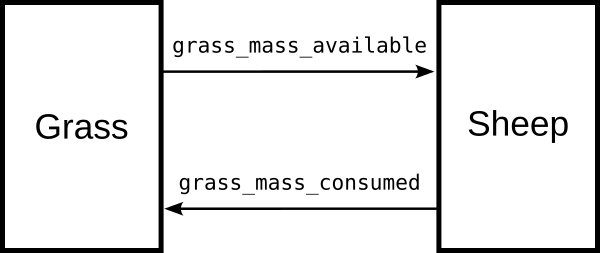
The Grass agents have a member variable mass, and an action GrassManager
which increases the mass logistically to a maximum value.
The mass can also be reduced, but only down to a minimum value.
The mass to be subtracted is read from a shared array “grass_mass_consumed”.
The GrassManager fills the shared array “grass_mass_available” with the
available grass mass in each cell (the available mass is equal to the
actual mass minus the minimum mass).
There should only be one grass agent in each cell.
The Sheep population uses the shared array “grass_mass_available” to create weighted random numbers for its WeightedMove action. Over time sheep lose mass (this is done by the Starver action), and are killed when thei masses is below a minimum mass. The population’s SheepManager action determines how much each sheep can eat in each cell, and accordingly fills the shared array “grass_mass_consumed”. The action AnimalReproducer is in charge of the sheep agents’ reproduction, which in this case is pathenogenetic (i.e. every agent can bear babies).
Population code:
Action code:
Contents
The Header Files (GrassPop)¶
The header file GrassPop.h defines an agent with an (additional) member m_dMass.
The class also contains two shared arrays
#ifndef __GRASSPOP_H__
#define __GRASSPOP_H__
#include <hdf5.h>
#include "SPopulation.h"
#include "GrassManager.h"
#define SHARE_GRASS_MASS_AVAILABLE "grass_mass_available"
#define SHARE_GRASS_MASS_CONSUMED "grass_mass_consumed"
struct GrassAgent : Agent {
double m_dMass;
};
class GrassPop : public SPopulation<GrassAgent> {
public:
GrassPop(SCellGrid *pCG, PopFinder *pPopFinder, int iLayerSize, IDGen **apIDG, uint32_t *aulState, uint *aiSeeds);
~GrassPop();
int addPopSpecificAgentData(int iAgentIndex, char **ppData);
void addPopSpecificAgentDataTypeQDF(hid_t *hAgentDataType);
int writeAdditionalDataQDF(hid_t hSpeciesGroup);
protected:
GrassManager<GrassAgent> *m_pGM;
//shared arrays
double *m_pdGrassAvailable;
double *m_pdGrassConsumed;
};
#endif
The Header File (SheepPop)¶
#ifndef __SHEEPPOP_H__
#define __SHEEPPOP_H__
#include <hdf5.h>
#include "SPopulation.h"
#include "WeightedMove.h"
#include "SingleEvaluator.h"
#include "MultiEvaluator.h"
#include "SheepManager.h"
#include "Starver.h"
#include "AnimalReproducer.h"
// here we just happen to know the share names
// perhaps we should store them in a singleton?
// names for shares created by GrassPop
#define SHARE_GRASS_MASS_AVAILABLE "grass_mass_available"
#define SHARE_GRASS_MASS_CONSUMED "grass_mass_consumed"
SheepPop expects the names of the two shared arrays to be “grass_mass_available” and “grass_mass_consumed”.
struct SheepAgent : Agent {
double m_dMass;
double m_dBabyMass;
};
Apart from the own mass, the mass of an agents newborn is specified.
class SheepPop : public SPopulation<SheepAgent> {
public:
SheepPop(SCellGrid *pCG, PopFinder *pPopFinder, int iLayerSize, IDGen **apIDG, uint32_t *aulState, uint *aiSeeds);
~SheepPop();
int preLoop();
int addPopSpecificAgentData(int iAgentIndex, char **ppData);
void addPopSpecificAgentDataTypeQDF(hid_t *hAgentDataType);
int makePopSpecificOffspring(int iAgent, int iMother, int iFather);
protected:
// pointers to the action obkects
SingleEvaluator<SheepAgent> *m_pSEGrass;
WeightedMove<SheepAgent> *m_pWM;
SheepManager<SheepAgent> *m_pSM;
Starver<SheepAgent> *m_pRS;
AnimalReproducer<SheepAgent> *m_pAR;
// the shared arrays
double *m_pdGrassMassAvailable;
double *m_pdGrassMassConsumed;
// the array of preferences: filled by ``m_pSEGrass`` and used by ``m_pWM''.
double *m_adPreferences;
};
#endif
The Implementation (GrassPop)¶
#include <stdio.h>
#include <omp.h>
#include <hdf5.h>
#include "SPopulation.cpp"
#include "LayerBuf.cpp"
#include "Prioritizer.cpp"
#include "Action.cpp"
#include "GrassManager.cpp"
#include "GrassPop.h"
The constructor mainly creates the GrassManager action and registers it, and creates and shares the two arrays.
//----------------------------------------------------------------------------
// constructor
//
GrassPop::GrassPop(SCellGrid *pCG, PopFinder *pPopFinder, int iLayerSize, IDGen **apIDG, uint32_t *aulState, uint *aiSeeds)
: SPopulation<GrassAgent>(pCG, pPopFinder, iLayerSize, apIDG, aulState, aiSeeds),
m_pGM(NULL),
m_pdGrassAvailable(NULL),
m_pdGrassConsumed(NULL) {
ArrayShare *pAS = ArrayShare::getInstance();
// create and share available grass mass array
m_pdGrassAvailable = new double[pCG->m_iNumCells];
int iResult = pAS->shareArray(SHARE_GRASS_MASS_AVAILABLE, pCG->m_iNumCells, m_pdGrassAvailable);
if (iResult != 0) {
printf("Couldnt share array [%p] with name [%s]\n", m_pdGrassAvailable, SHARE_GRASS_MASS_AVAILABLE);
} else {
printf("GrassPop shared [%p] as [%s]\n", m_pdGrassAvailable, SHARE_GRASS_MASS_AVAILABLE);
}
// create and share consumed grass array
m_pdGrassConsumed = new double[pCG->m_iNumCells];
iResult = pAS->shareArray(SHARE_GRASS_MASS_CONSUMED, pCG->m_iNumCells, m_pdGrassConsumed);
if (iResult != 0) {
printf("Couldnt share array [%p] with name [%s]\n", m_pdGrassConsumed, SHARE_GRASS_MASS_CONSUMED);
} else {
printf("GrassPop shared [%p] as [%s]\n", m_pdGrassConsumed, SHARE_GRASS_MASS_CONSUMED);
}
// we only have one action
m_pGM = new GrassManager<GrassAgent>(this, m_pCG, "", SHARE_GRASS_MASS_AVAILABLE, SHARE_GRASS_MASS_CONSUMED);
m_prio.addAction(m_pGM);
}
The destructor deletes the GrassManager action and the shared arrays.
//----------------------------------------------------------------------------
// destructor
// delete the allocated arrays
//
GrassPop::~GrassPop() {
if (m_pGM != NULL) {
delete m_pGM;
}
if (m_pdGrassAvailable != NULL) {
delete[] m_pdGrassAvailable;
}
if (m_pdGrassConsumed != NULL) {
delete[] m_pdGrassConsumed;
}
ArrayShare::freeInstance();
}
//----------------------------------------------------------------------------
// addPopSpecificAgentData
// read additional data (m_dMass) from pop file
//
int GrassPop::addPopSpecificAgentData(int iAgentIndex, char **ppData) {
int iResult = 0;
iResult = addAgentDataSingle(ppData, &m_aAgents[iAgentIndex].m_dMass);
return iResult;
}
//----------------------------------------------------------------------------
// addPopSpecificAgentDataTypeQDF
// extend the agent data type to include additional data in the output
//
void GrassPop::addPopSpecificAgentDataTypeQDF(hid_t *hAgentDataType) {
GrassAgent ga;
H5Tinsert(*hAgentDataType, "Mass", qoffsetof(ga, m_dMass), H5T_NATIVE_DOUBLE);
}
The GrassPop writes the array of available grass mass to file as an additional array.
//----------------------------------------------------------------------------
// writeAdditionalDataQDF
// write the available grass mass as an array in the population group
//
int GrassPop::writeAdditionalDataQDF(hid_t hSpeciesGroup) {
xha_printf("GrassPop::writeAdditionalDataQDF() reached\n");
int iResult = qdf_writeArray(hSpeciesGroup, "GrassMassAvail", m_pCG->m_iNumCells, m_pdGrassAvailable, H5T_NATIVE_DOUBLE);
if (iResult != 0) {
LOG_ERROR("[GrassPop::writeAdditionalDataQDF] couldn't write GrassMassAvail data");
}
return iResult;
}
The Implementation (SheepPop)¶
#include <stdio.h>
#include <omp.h>
#include <hdf5.h>
#include "SPopulation.cpp"
#include "LayerBuf.cpp"
#include "Prioritizer.cpp"
#include "Action.cpp"
#include "SheepManager.cpp"
#include "Starver.cpp"
#include "WeightedMove.cpp"
#include "SingleEvaluator.cpp"
#include "MultiEvaluator.cpp"
#include "AnimalReproducer.cpp"
#include "SheepPop.h"
In its constructor SheepPop allocates an array for cell preferences based on available grass mass which will be filled by the SingleEvaluator
object and used by the WeightedMove object. This array is conceptually an array of 7-tuples, with one tuple for each cell.
The first element of such a tuple contains the preference value for the cell to which the tuple belongs.
The following values contain the preference values for the neighboring cells.

Then the action objects (SingleEvaluator, WeightedMove, SheepManager, Starver, and AnimalReproducer are created and registered with the Prioritizer.
//----------------------------------------------------------------------------
// constructor
//
SheepPop::SheepPop(SCellGrid *pCG, PopFinder *pPopFinder, int iLayerSize, IDGen **apIDG, uint32_t *aulState, uint *aiSeeds)
: SPopulation<SheepAgent>(pCG, pPopFinder, iLayerSize, apIDG, aulState, aiSeeds),
m_pSEGrass(NULL),
m_pWM(NULL),
m_pSM(NULL),
m_pSS(NULL),
m_pAR(NULL),
m_pdGrassMassAvailable(NULL),
m_pdGrassMassConsumed(NULL),
m_adPreferences(NULL) {
int iMaxNeighbors = this->m_pCG->m_iConnectivity;
int iArrSize = this->m_pCG->m_iNumCells * (iMaxNeighbors + 1);
m_adPreferences = new double[iArrSize];
memset(m_adPreferences, 0, pCG->m_iNumCells*sizeof(double));
m_pSEGrass = new SingleEvaluator<SheepAgent>(this, m_pCG, "Grass", m_adPreferences, (double*)m_pdGrassMassAvailable, "", true, EVENT_ID_NONE, true);
m_pWM = new WeightedMove<SheepAgent>(this, m_pCG, "", m_apWELL, m_adPreferences);
m_pSM = new SheepManager<SheepAgent>(this, m_pCG, "",
SHARE_GRASS_MASS_AVAILABLE, SHARE_GRASS_MASS_CONSUMED);
m_pSS = new Starver<SheepAgent>(this, m_pCG, "");
m_pAR = new AnimalReproducer<SheepAgent>(this, m_pCG, "", m_apWELL);
m_prio.addAction(m_pSEGrass);
m_prio.addAction(m_pWM);
m_prio.addAction(m_pSM);
m_prio.addAction(m_pSS);
m_prio.addAction(m_pAR);
}
In the destructor the array allocated in the constructor as well as all action objects are deleted.
//----------------------------------------------------------------------------
// destructor
//
SheepPop::~SheepPop() {
if (m_adPreferences != NULL) {
delete[] m_adPreferences;
}
if (m_pSEGrass != NULL) {
delete m_pSEGrass;
}
if (m_pWM != NULL) {
delete m_pWM;
}
if (m_pSM != NULL) {
delete m_pRM;
}
if (m_pSS != NULL) {
delete m_pRS;
}
if (m_pAR != NULL) {
delete m_pAR;
}
ArrayShare::freeInstance();
}
In the method preLoop the array of available grass mass, which was created by GrassPop, is retrieved and passed to the SingleEvaluator.
At the time of the constructor there is no guarantee, that the array has already been shared, so we do this here.
//----------------------------------------------------------------------------
// preLoop
// read additional data from pop file
//
int SheepPop::preLoop() {
int iResult = 0;
m_pdGrassMassAvailable = (double *) ArrayShare::getInstance()->getArray(SHARE_GRASS_MASS_AVAILABLE);
if (m_pdGrassMassAvailable == NULL) {
iResult = -1;
} else {
m_pSEGrass->setInputData(m_pdGrassMassAvailable);
iResult = SPopulation<SheepAgent>::preLoop();
}
return iResult;
}
//----------------------------------------------------------------------------
// addPopSpecificAgentData
// read additional data from pop file
//
int SheepPop::addPopSpecificAgentData(int iAgentIndex, char **ppData) {
int iResult = 0;
iResult = addAgentDataSingle(ppData, &m_aAgents[iAgentIndex].m_dMass);
m_aAgents[iAgentIndex].m_dBabyMass = 0;
return iResult;
}
//----------------------------------------------------------------------------
// addPopSpecificAgentDataTypeQDF
// extend the agent data type to include additional data in the output
//
void SheepPop::addPopSpecificAgentDataTypeQDF(hid_t *hAgentDataType) {
SheepAgent ra;
H5Tinsert(*hAgentDataType, "Mass", qoffsetof(ra, m_dMass), H5T_NATIVE_DOUBLE);
}
When a baby is born we set its initial mass and subtract this from the parent’s mass.
//----------------------------------------------------------------------------
// makePopSpecificOffspring
//
int SheepPop::makePopSpecificOffspring(int iAgent, int iMother, int iFather) {
//int iThread = omp_get_thread_num();
//double fM = 31 + this->m_apWELL[iThread]->wrandr(0.0,1.0);
float fM = m_aAgents[iMother].m_dBabyMass;
m_aAgents[iAgent].m_dMass = fM;
m_aAgents[iMother].m_dMass -= fM;
// m_aAgents[iAgent].m_iMateIndex = -3;
//printf("birth: M %d (%f), B %d (%f)\n", iMother, m_aAgents[iMother].m_dMass, iAgent, m_aAgents[iAgent].m_dMass);
return 0;
}
XML and DAT¶
The xml file for “GrassPop” is simple:
<class name="GrassPop" species_name="grass" species_id="104">
<module name="GrassManager">
<param name="MinMass" value="1.0"/>
<param name="MaxMass" value="10.0"/>
<param name="GrowthRate" value="0.008"/>
</module>
<priorities>
<prio name="GrassManager" value="1"/>
</priorities>
</class>
Since we want grass to grow on every cell, and there should be only one grass agent per cell, the dat file for GrassPop contains 250’000 entries (one for each cell in the 500x5000 grid):
#(NodeID);LifeState;AgentID;BirthTime;Gender;Age
(0);1;0;-10;0;5
(1);1;1;-10;0;5
(2);1;2;-10;0;5
...
The xml for SheepPop must provide values for all action attributes:
<class name="SheepPop" species_name="sheep" species_id="105">
<module name="WeightedMove">
<param name="WeightedMove_prob" value="0.1"/>
</module>
<module name="SingleEvaluator" id="Grass" >
</module>
<module name="Starver">
<param name="StarveMass" value="0.4"/>
<param name="MassDecay" value="0.1"/>
</module>
<module name="AnimalReproducer">
<param name="MassFert" value="6.0"/>
<param name="MassBaby" value="3.0"/>
<param name="BirthProb" value="0.2"/>
</module>
<priorities>
<prio name="SheepManager" value="2"/>
<prio name="AnimalReproducer" value="4"/>
<prio name="SingleEvaluator[Grass]" value="5"/>
<prio name="WeightedMove" value="6"/>
<prio name="Starver" value="7"/>
</priorities>
</class>
In the dat file for SheepPop there is only one agent placed in the middle of the field:
#(NodeID);LifeState;AgentID;BirthTime;Gender;Age
(125250);1;0;-10.0;0;8
Grid¶
For this example we use flat 500x500 geometry with torus identifications and uniform altitude (250’000 cells).
Compiling and Running¶
Assuming that you have created the tutorial directories with the script build_tut_dirs.py,
lets first create alternative population and action directories with the script build_alt_modpop.py:
${QHG4_DIR}/useful_stuff/build_alt_modpop.py tut_08 GrassPop SheepPop
This builds alternative population directories tut_08_pop and tut_08_mod with the files needed to compile GrassPop and SheepPop``.
To compile, change to the QHG4 root directory and type:
OPT=1 SHORT=tut_08 make clean QHGMain
Here we set the environemnt variable OPT to get optimized compilation (see Compiling QHG)
Now cd to the tutorial subdirectory tutorial_08 and start QHGMain
${QHG4_DIR}/app/QHGMain --read-config=tutorial_08.cfg > tutorial_08.out
This tells QHGMain to read the parameters from the file tutorial_08.cfg and write all output to the file tutorial_08.out (this output contains more information than the logfile)
The config file tutorial_08.cfg was created when the tutorial directories were created with build_tut_dirs.py.
--data-dirs=${QHG4_DIR}/tutorial_data/grids/
--events='write|grid+geo+pop:grass~+pop:sheep#@10'
--grid=torus_500x500.qdf
--log-file=tutorial_08.log
--num-iters=5000
--output-dir=output/tutorial_08
--output-prefix=tut
--pops='tut_Grass.xml:tut_Grass.dat,tut_Sheep.xml:tut_Sheep.dat'
--shuffle=92244
--start-time=0
data-dirsA list of directories to search for data.
eventsA list of events (see Events). Here we write output files every 10 steps, containing the additional data arrays of GrassPop (in this case the array of available grassmass) but not the agent data, and for SheepPop only the agent data is written.
gridPath to the grid on which to run the simulation, in this case the 500x500 torus.
log-fileLog file for ‘high level’ messages.
num-itersNumber of iterations for the simulation (5000 steps)
output-dirOutput directory for qdf files.
output-prefixA prefix prepended to the names of all output files
popsThe files for population attributes (
tut_Grass_500x500.xml,tut_Sheep_500x500.xml) and agent attributes (tut_SexualPop.dat). These files were created bybuild_tut_dirs.py.shuffleA kiind of seed for the random nummber generators. Same
shufflevalues result in identical simulations.start-timeThe ‘real’ time corresponding to step 0.
Visualizing the Results¶
A simulation with the attributes described above was run for 10’000 steps, starting with a single sheep agent in the center of the field. Initially each grass agent had half of the maximum mass.
After every 10 time steps a snapshot of the simulation state was made. These snapshots were visualized in VisIt as a group, with all annotations removed (“Control|Annotations”), and then turned into a video using VisIt’s “Save as Movie…” command in the “File” menu.
The colors represent the maximum available grass mass in each cell (blue:0, red max).
Each black dot represents a single sheep agent.