QHG Tutorial 07 - Environment interaction II¶
In this chapter we extend our example by making the agents aware of two arrays of the environment, the carrying capcity calculated from the Net Primary Production (NPP) and altitude. Given an NPP value, a local carrying is calculated by means of an NPPCapacity object.
To do this, we combine two instances of SingleEvaluator, one for NPP and one for altitude, by means of a MultiEvaluator.
The output of the MultiEvaluator will now be the input for the WeightedMove object.
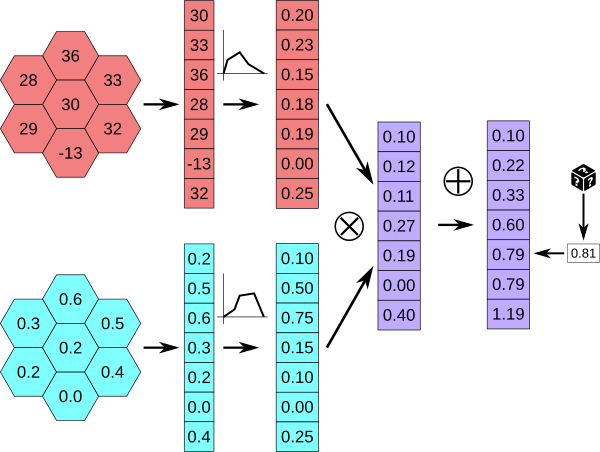
The environment values (for altitude and NPP are each copied to an array, to which poly line functions are applied.
The resulting arrays are combined by the MultiEvaluator (here the corresponding values are simply multiplied), cumulated, and used as a lookup table to create a weighted random number.
In this example a uniform random number R between 0 and the highest number in the cumulated array is drawn, and the index of the smallest cumulated value greater than R is chosen.
Here the random number 0.81 is less than the value at index 4, 0.95, so the chosen index is 4.
Those indexes are selected with a probability proportional to the preference values of he corresponding cells¶
Population code:
Action code:
The Header File¶
The include file shows that Verhulst was replaced by VerhulstVarK, and that MultiEvaluator and NPPCapacity are additionally included.
The agent structure is equal to the one in the previous example.
#ifndef __TUT_ENVIRONCAPALTPOP_H__
#define __TUT_ENVIRONCAPALTPOP_H__
#include "GetOld.h"
#include "OldAgeDeath.h"
#include "Fertility.h"
#include "VerhulstVarK.h"
#include "RandomPair.h"
#include "SingleEvaluator.h"
#include "WeightedMove.h"
#include "MultiEvaluator.h"
#include "NPPCapacity.h"
struct tut_EnvironCapAltAgent : Agent {
float m_fAge;
float m_fLastBirth;
int m_iMateIndex;
};
The class now has a pointer to VerhulstVarK instead of Verhulst, a pointer to MultiEvaluator instead of SingleEvaluator,
and an additional pointer to NPPCapacity.
class tut_EnvironCapAltPop : public SPopulation<tut_EnvironCapAltAgent> {
public:
tut_EnvironCapAltPop(SCellGrid *pCG, PopFinder *pPopFinder, int iLayerSize, IDGen **apIDG, uint32_t *aulState, uint *aiSeeds);
~tut_EnvironCapAltPop();
int makePopSpecificOffspring(int iAgent, int iMother, int iFather);
int addPopSpecificAgentData(int iAgentIndex, char **ppData);
void addPopSpecificAgentDataTypeQDF(hid_t *hAgentDataType);
virtual int updateEvent(int EventID, char *pData, float fT);
virtual void flushEvents(float fT);
protected:
WeightedMove<tut_EnvironCapAltAgent> *m_pWM;
MultiEvaluator<tut_EnvironCapAltAgent> *m_pME;
VerhulstVarK<tut_EnvironCapAltAgent> *m_pVerVarK;
RandomPair<tut_EnvironCapAltAgent> *m_pPair;
GetOld<tut_EnvironCapAltAgent> *m_pGO;
OldAgeDeath<tut_EnvironCapAltAgent> *m_pOAD;
Fertility<tut_EnvironCapAltAgent> *m_pFert;
NPPCapacity<tut_EnvironCapAltAgent> *m_pNPPCap;
double *m_adEnvWeights;
double *m_adCapacities;
bool m_bUpdateNeeded;
};
The member variables reflect the new situation of required actions. The member functions are identical to the ones discussed in the prevouis example.
Code: tut_EnvironCapAltPop.h
The Implementation¶
The include files, too, reflect the changes in the actions:
#include <omp.h>
#include <hdf5.h>
#include "EventConsts.h"
#include "SPopulation.cpp"
#include "Prioritizer.cpp"
#include "LayerBuf.cpp"
#include "LayerArrBuf.cpp"
#include "Action.cpp"
#include "GetOld.cpp"
#include "OldAgeDeath.cpp"
#include "Fertility.cpp"
#include "VerhulstVarK.cpp"
#include "RandomPair.cpp"
#include "WeightedMove.cpp"
#include "SingleEvaluator.cpp"
#include "MultiEvaluator.cpp"
#include "NPPCapacity.cpp"
#include "tut_EnvironCapAltPop.h"
The constructor first allocates the arrays for local capacities and for altitudes,
which are needed for communication between the evaluators and other objects.
Then the SingleEvaluator objects for altitude and capacity created and assigned to the MultiEvaluator object.
After all action objects have been created, they are registered to the priority manager.
During the simulation the altitude evaluator will read from the geography’s altitude array, whenever it has changed. On the other hand the NPPCapacity object will
calculate the local carrying capacity for each cell and insert them into the array m_adCapacities.
The MultiEvaluator wil combine the results of its two single evaluators and returns the result in the array m_adEnvWeights,
which in turn is read by the WeightedMove object to move an agent in a weighted random direction (see image above).
tut_EnvironCapAltPop::tut_EnvironCapAltPop(SCellGrid *pCG, PopFinder *pPopFinder, int iLayerSize, IDGen **apIDG, uint32_t *aulState, uint *aiSeeds)
: SPopulation<tut_EnvironCapAltAgent>(pCG, pPopFinder, iLayerSize, apIDG, aulState, aiSeeds) {
int iCapacityStride = 1;
m_adCapacities = new double[m_pCG->m_iNumCells];
memset(m_adCapacities, 0, m_pCG->m_iNumCells * sizeof(double));
m_adEnvWeights = new double[m_pCG->m_iNumCells * (m_pCG->m_iConnectivity + 1)];
memset(m_adEnvWeights, 0, m_pCG->m_iNumCells * (m_pCG->m_iConnectivity + 1)*sizeof(double));
// prepare parameters for MultiEvaluator, which will compute actual carrying capacity
MultiEvaluator<tut_EnvironCapAltAgent>::evaluatorinfos mEvalInfo;
// add altitude evaluation - output is NULL because it will be set by MultiEvaluator
SingleEvaluator<tut_EnvironCapAltAgent> *pSEAlt = new SingleEvaluator<tut_EnvironCapAltAgent>(this, m_pCG, "Alt", NULL, (double*)m_pCG->m_pGeography->m_adAltitude, "AltPref", true, EVENT_ID_GEO);
mEvalInfo.push_back(std::pair<std::string, Evaluator<tut_EnvironCapAltAgent>*>("Multi_weight_alt", pSEAlt));
// add NPP evaluation - output is NULL because it will be set by MultiEvaluator
SingleEvaluator<tut_EnvironCapAltAgent> *pSECap = new SingleEvaluator<tut_EnvironCapAltAgent>(this, m_pCG, "NPP", NULL, m_adCapacities, "", true, EVENT_ID_VEG);
mEvalInfo.push_back(std::pair<std::string, Evaluator<tut_EnvironCapAltAgent>*>("Multi_weight_npp", pSECap));
m_pME = new MultiEvaluator<tut_EnvironCapAltAgent>(this, m_pCG, "NPP+Alt", m_adEnvWeights, mEvalInfo, MODE_ADD_SIMPLE, true); // true: delete evaluators
m_pNPPCap = new NPPCapacity<tut_EnvironCapAltAgent>(this, m_pCG, "", m_apWELL, m_adCapacities, iCapacityStride);
m_pWM = new WeightedMove<tut_EnvironCapAltAgent>(this, m_pCG, "", m_apWELL, m_adEnvWeights);
m_pGO = new GetOld<tut_EnvironCapAltAgent>(this, m_pCG, "");
m_pOAD = new OldAgeDeath<tut_EnvironCapAltAgent>(this, m_pCG, "", m_apWELL);
m_pFert = new Fertility<tut_EnvironCapAltAgent>(this, m_pCG, "");
m_pVerVarK = new VerhulstVarK<tut_EnvironCapAltAgent>(this, m_pCG, "", m_apWELL, m_adCapacities, iCapacityStride);
m_pPair = new RandomPair<tut_EnvironCapAltAgent>(this, m_pCG, "", m_apWELL);
// adding all actions to prioritizer
m_prio.addAction(m_pGO);
m_prio.addAction(m_pOAD);
m_prio.addAction(m_pFert);
m_prio.addAction(m_pVerVarK);
m_prio.addAction(m_pPair);
m_prio.addAction(m_pME);
m_prio.addAction(m_pWM);
m_prio.addAction(m_pNPPCap);
}
The destructor deletes all action objects created by the constructor, as well as the communication array.
//----------------------------------------------------------------------------
// destructor
//
tut_EnvironCapAltPop::~tut_EnvironCapAltPop() {
if (m_pGO != NULL) {
delete m_pGO;
}
if (m_pOAD != NULL) {
delete m_pOAD;
}
if (m_pFert != NULL) {
delete m_pFert;
}
if (m_pVerVarK != NULL) {
delete m_pVerVarK;
}
if (m_pPair != NULL) {
delete m_pPair;
}
if (m_pME != NULL) {
delete m_pME;
}
if (m_pWM != NULL) {
delete m_pWM;
}
if (m_pNPPCap != NULL) {
delete m_pNPPCap;
}
if (m_adEnvWeights != NULL) {
delete[] m_adEnvWeights;
}
if (m_adCapacities != NULL) {
delete[] m_adCapacities;
}
}
The methods addPopSpecificAgentData() and addPopSpecificAgentDataTypeQDF() are practically identical to the ones in the previous chapter, so there’s no need to look at them in more detail.
The method makePopSpecificOffspring() is also identical to the one in the previous examples.
//----------------------------------------------------------------------------
// makePopSpecificOffspring
//
int tut_SexualPop::makePopSpecificOffspring(int iAgent, int iMother, int iFather) {
int iResult = 0;
m_aAgents[iAgent].m_fAge = 0.0;
m_aAgents[iAgent].m_fLastBirth = 0.0;
m_aAgents[iAgent].m_iMateIndex = -3;
// SPopulation assigns random genders to a baby, ehich is ok here
return iResult;
}
The mate index is set to a negative number to denote that this agent is not paired (yet).
The method updateEvents() is called when an event is fired.
//----------------------------------------------------------------------------
// updateEvent
// react to events
// EVENT_ID_GEO : kill agent if under ice or water
//
int tut_EnvironAltPop::updateEvent(int iEventID, char *pData, float fT) {
if (iEventID == EVENT_ID_GEO) {
// drown
int iFirstAgent = getFirstAgentIndex();
if (iFirstAgent != LBController::NIL) {
int iLastAgent = getLastAgentIndex();
#pragma omp parallel for
for (int iAgent = iFirstAgent; iAgent <= iLastAgent; iAgent++) {
if (m_aAgents[iAgent].m_iLifeState > LIFE_STATE_DEAD) {
int iCellIndex = m_aAgents[iAgent].m_iCellIndex;
if ((this->m_pCG->m_pGeography->m_adAltitude[iCellIndex] < 0) ||
(this->m_pCG->m_pGeography->m_abIce[iCellIndex] > 0)) {
registerDeath(iCellIndex, iAgent);
}
}
}
}
// make sure they are removed before step starts
if (m_bRecycleDeadSpace) {
recycleDeadSpaceNew();
} else {
performDeaths();
}
// update counts
updateTotal();
updateNumAgentsPerCell();
// clear lists to avoid double deletion
initListIdx();
}
notifyObservers(iEventID, pData);
return 0;
}
Here we only react if a change happened to the geography (usually when a new QDF file is loaded). In this case we check all agents in a parallelized loop adding those under water (negative altitude) and those under ice to the death list Subsequently, the deaths are actually performed, and several counts are updated.
Code: tut_EnvironCapAltPop.h
XML and DAT¶
In the xml file for this example a new module for NPPCpacity, and one for MultiEvaluator have been added.
The submodules for the SingleEvaluator objects show that the altitudes are processed by a poly line object, whereas the NPP vlaues are used “as is”.
<class name="tut_EnvironCapAltPop" species_name="sapiens" species_id="104">
<module name="OldAgeDeath">
<param name="OAD_max_age" value="60.0"/>
<param name="OAD_uncertainty" value="0.1"/>
</module>
<module name="WeightedMove">
<param name="WeightedMove_prob" value="0.07"/>
</module>
<module name="Fertility">
<param name="Fertility_interbirth" value="2.0"/>
<param name="Fertility_max_age" value="50.0"/>
<param name="Fertility_min_age" value="15.0"/>
</module>
<module name="NPPCapacity">
<param name="NPPCap_K_max" value="38.4716796875"/>
<param name="NPPCap_K_min" value="0.0"/>
<param name="NPPCap_NPP_max" value="1.0576171875"/>
<param name="NPPCap_NPP_min" value="0.0"/>
<param name="NPPCap_coastal_factor" value="0.4"/>
<param name="NPPCap_coastal_max_latitude" value="66.0"/>
<param name="NPPCap_coastal_min_latitude" value="50.0"/>
<param name="NPPCap_water_factor" value="0.5349609375"/>
<param name="NPPCap_efficiency" value="1"/>
</module>
<module name="MultiEvaluator" id="NPP+Alt" >
<param name="Multi_weight_alt" value="0.2" />
<param name="Multi_weight_npp" value="0.8" />
<module name="SingleEvaluator" id="Alt" >
<param name="AltPref" value="-0.1 0 0.1 0.01 1500 1.0 2000 1 3000 -9999" />
</module>
<module name="SingleEvaluator" id="NPP" >
</module>
</module>
<module name="VerhulstVarK">
<param name="Verhulst_b0" value="0.8" />
<param name="Verhulst_d0" value="0.001" />
<param name="Verhulst_theta" value="0.1" />
</module>
<priorities>
<prio name="NPPCapacity" value="1"/>
<prio name="GetOld" value="2"/>
<prio name="OldAgeDeath" value="3"/>
<prio name="WeightedMove" value="4"/>
<prio name="MultiEvaluator[NPP+Alt]" value="5"/>
<prio name="Fertility" value="6"/>
<prio name="RandomPair" value="7"/>
<prio name="VerhulstVarK" value="8"/>
</priorities>
</class>
The DAT file is equal to the one in the previous example, all agent start in South Africa.
#Longitude;Latitude;LifeState;AgentID;BirthTime;Gender;Age;LastBirth
28.0<46;-26.044;1; 1;-10.0;0;10.0;-1.0
28.046;-26.044;1; 2;-10.0;1;10.0;-1.0
28.046;-26.044;1; 3;-10.0;0;10.0;-1.0
28.046;-26.044;1; 4;-10.0;1;10.0;-1.0
...
28.046;-26.044;1;19;-10.0;0;10.0;-1.0
28.046;-26.044;1;20;-10.0;1;10.0;-1.0
Grid¶
For this example we use a subdivided icosahedron with earths’s geography: there are oceans and continents.
Compiling and Running¶
Again, we assume that you have created the tutorial directories with the script build_tut_dirs.py.
First lets create alternative population and action directories with the script build_alt_modpop.py:
${QHG4_DIR}/useful_stuff/build_alt_modpop.py tut_07 tut_EnvironCapAltPop
This builds alternative population directories tut_07_pop and tut_07_mod with the files needed to compile tut_EnvironAltPop.
To compile, change to the QHG4 root directory and type:
OPT=1 SHORT=tut_07 make clean QHGMain
Here we set the environemnt variable OPT to get optimized compilation (see Compiling QHG)
Now cd to the tutorial subdirectory tutorial_07 and start QHGMain
${QHG4_DIR}/app/QHGMain --read-config=tutorial_07.cfg > tutorial_07.out
This tells QHGMain to read the parameters from the file tutorial_07.cfg and write all output to the file tutorial_07.out (this output contains more information than the logfile)
The config file tutorial_07.cfg was created when the tutorial directories were created with build_tut_dirs.py.
--data-dirs=${QHG4_DIR}/tutorial_data/grids/
--events='write|grid+geo+pop:sapiens@1000'
--grid=eq64Alt.qdf
--log-file=tutorial_07.log
--num-iters=80000
--output-dir=output/tutorial_07
--output-prefix=tut
--pops='tut_EnvironCapAlt.xml:tut_EnvironCapAlt.dat'
--shuffle=92244
--start-time=-80000
data-dirsA list of directories to search for data (not relevant for the OldAgeDie populations).
eventsA list of events (see Events). Here we write output files containing grid, geography and agent data every 1000 steps.
gridPath to the grid on which to run the simulation.
log-fileLog file for ‘high level’ messages.
num-itersNumber of iterations for the simulation (10000 steps)
output-dirOutput directory for qdf files.
output-prefixA prefix prepended to the names of all output files
popsThe files for population attributes (
tut_SexualPop.xml) and agent attributes (tut_SexualPop.dat). These files were created bybuild_tut_dirs.py.shuffleA kind of seed for the random number generators. Same
shufflevalues result in identical simulations.start-timeThe ‘real’ time corresponding to step 0.